Identification of microorganisms in pure and mixed cultures: 16S rRNA sequencing and metagenomics
We provide laboratory contract research services to identify classified and unclassified microorganisms from samples containing pure cultures and mixed prokaryotic and eukaryotic populations. Using highly selective PCR primer sets we identify all organisms in the sample or organisms in selective groups (i.e. prokaryotic vs. eukaryotic). The process uses two-step PCR amplification to generate sequencing libraries. Paired-end sequencing performed on the MiSeq System (Illumina) and analyzed using the QIIME pipeline.
Example of a 16S rRNA metagenomics experiment using primer sets for amplifying the V1/V2 ribosomal RNA region. First round PCR using primers (underlined nucleotides represent 16S rRNA sequences:
Hyb8F_rRNA: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTTTGATCMTGGCTCAG -3′
Hyb338R_rRNA: 3′-TGAGGATGCCCTCCGTGACAGAGAATATGTGTAGAGGCTCGGGTGCTCTG-5′
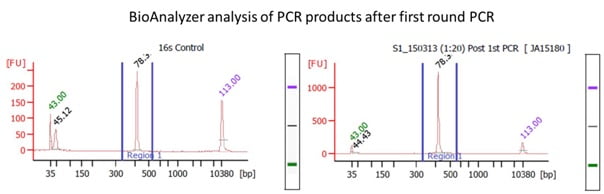
The example above uses primers to amplify and sequence the V1/V2 variable regions of the 16S rRNA gene. We also provide service for sequencing the V4 region.
Genotyping applications largely depend on sequencing data (most often 16S or 5S). Altogen Labs performs extraction and purification of genomic DNA, 16s sequencing (using Altogen proprietary library of 48 different primer sets), and produces analyzed results. Depending on experimental assay design, sequencing can be used to discriminate microbial species, types, and strains, or detect genetic mutations that confer resistance to antibiotics or antiviral drugs.
We can provide an immediate price quote (contact e-mail: info@altogenlabs.com). Please note that experimental details will help us provide an accurate quote and timeline estimate.

